RNase H 活性的缺失有利于其应用,因为在长孵育时间的时候,RNase H 会开始降解模板。虽然许多研究者已经成功使用M-MLV RT(H+) 进行了一些cDNA 制备应用及分析,但是缺失了RNase H 活性的逆转录酶为长cDNA 的制备及包含高比例全长cDNA 的文库制备提供了更好的选择。
特点 - 优点
• 无RNase H 活性:为以长RNA 为模板制备全长cDNA 提供了优化条件。
• 温度稳定性:点突变产物的热稳定性可防止与二级结构相关的问题。
• 聚合酶活性增强:M-MLV RT [H-], Point Mutant (Cat.# M3681)与缺失突变得到的M-MLV RT(Cat.# M5301) 相比,具有更高的cDNA 产率。
• 提供5× 反应缓冲液:250mM Tris-HCl (pH 8.3,25℃ ),375mM KCl,15mM MgCl2,50mM DTT。
• 广泛的工作范围:提高了逆转录的性能,使得可以在更广泛的酶浓度和底物浓度的条件下工作。
应用
• cDNA第一链合成
• 引物延伸
来源
Recombinant E. coli strain.
存储缓冲液
20mM Tris-HCl (pH 7.5 at 25°C), 200mM NaCl, 0.1mM EDTA, 1mM DTT, 0.01% Nonidet® P-40 and 50% glycerol.
储存条件
储存于-20℃。
单位定义
One unit is the amount of enzyme required to incorporate 1nmol of deoxynucleotide into acid-precipitable material in 10 minutes at 37°C. The reaction conditions are: 50mM Tris-HCl (pH 8.3 at 25°C), 7mM MgCl2, 40mM KCl, 10mM DTT, 0.1mg/ml BSA, 0.5mM [3H]dTTP, 0.025mM oligo(dT), 0.25mM poly(A) and 0.01% NP-40.
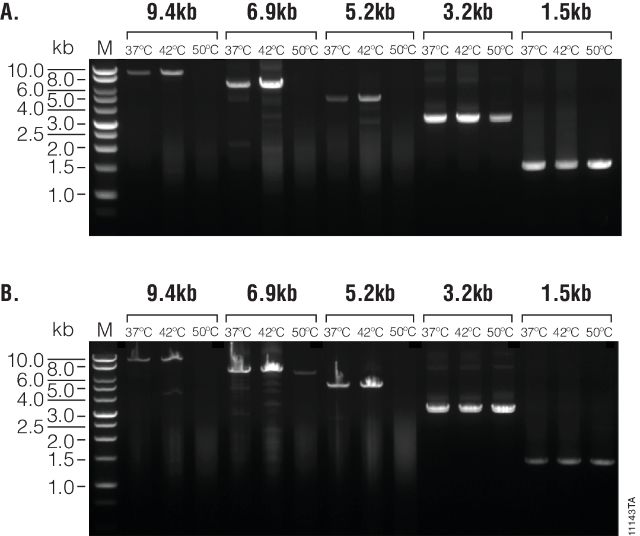
Figure 1. Thermal stability and synthesis of large transcripts.
cDNAs were synthesized from 1µg of total human RNA using M-MLV (H–), Point Mutant, (Panel A) or SuperScript® II (Panel B) reverse transcriptase at 37°C, 42°C and 50°C. GoTaq® Long PCR amplifications were performed on each transcription reaction using five primer sets, amplifying products from 9.4kb to 1.5kb. Ten microliters of the 9.4, 6.9, and 5.2kb PCR products and 1µl of the 3.2 and 1.5kb PCR products were separated on a 1% agarose gel. Lane M. BenchTop 1kb DNA Ladder (Cat.# G7541).
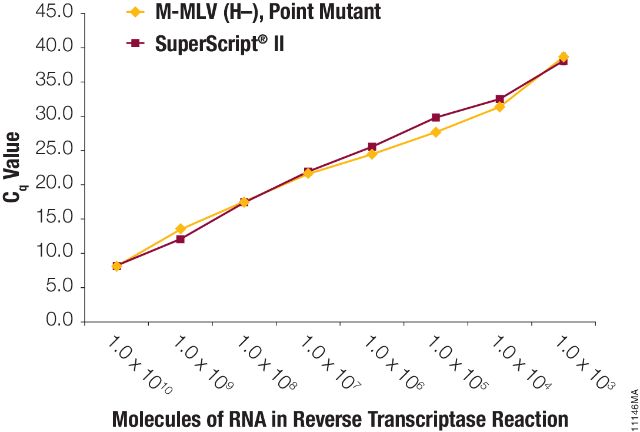
Figure 2. Sensitivity in a qPCR amplification.
Kanamycin control RNA was serially diluted tenfold (1 × 1010 to 1 × 101), spiked into 1ng of total human RNA and used as template in reverse transcription reactions with M-MLV (H–), Point Mutant, (Panel A) or SuperScript®II (Panel B) reverse transcriptase. Quantitative PCR amplification was performed on 5µl of the GoTaq® qPCR System with kanamycin control primers. Cq values are plotted versus molecules of RNA in the reverse transcription reactions. Cq values could not be determined at amounts less than 1 × 103 molecules. N=3.
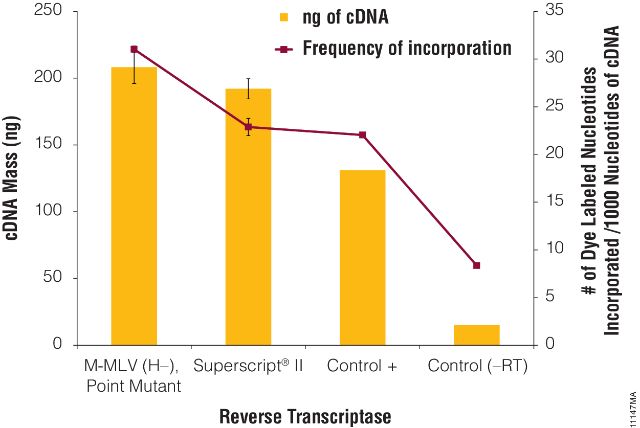
Figure 3. Label incorporation.
Reverse transcription reactions were performed as described in the ChipShot™ Direct Labeling and Clean-Up System Technical Manual, #TM286, using the positive control transcriptase in the kit and M-MLV (H–), Point Mutant, or SuperScript® II reverse transcriptase. A no-reverse transcriptase reaction (–RT) was also performed as a negative control. Reactions were cleaned-up using the ChipShot™ Direct Labeling and Clean-Up System (Cat.# Z4100). The amount of cDNA generated and dye incorporated were determined using A260 and A550 values. N=2.